Gene Expression
Gene expression is the process whereby the information encoded in the DNA of a gene is converted into a protein, which confers the observable phenotype upon the cell.
Simply defined, a gene Opens in new window is the nucleic acid sequence that is necessary for the synthesis of a functional peptide or protein in a temporal and tissue-specific manner. However, a gene is not directly translated into a protein; it is expressed via a nucleic acid intermediary called messenger RNA (mRNA).
The transcriptional unit of every gene is the sequence of DNA transcribed into a single mRNA molecue, starting at the promoter and ending at the terminator regions. The essential features of a gene and mRNA are presented in Figure X-1.
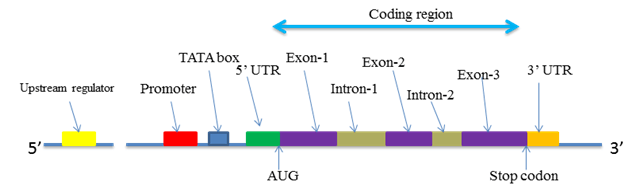
|
| Figure X-1 | Essential features of the gene |
The DNA sequence of a gene comprises two non-coding (or untranslated) regions at the beginning and end of the gene coding region. The non-coding promoter and terminator regions of the DNA are partially transcribed, but not translated and therefore form the 5’ and 3’ untranslated regions (UTR) of mRNA.
Although the non-coding regions of a gene and mRNA are not translated into the protein product of the gene, they contain critical parts of the genetic information involved in regulation of gene expression and the characteristics of the protein production.
The promoter region is located immediately upstream of the gene coding region; it contains DNA sequences, known as the TATA and CAAT boxes, which define the DNA binding sites at which transcription starts and regulate the rate of gene expression.
The TATA box is an AT-rich sequence that occurs about 30bp (–30bp) upstream from the transcriptional start site. The CAAT box contains this short DNA sequence about 80bp upstream (–80bp) of the start site. These sequences, together with binding sites for other transcription factors, regulate the rate of tissue-specific gene expression.
Transcription starts at the CAP site, so-called because following transcription the 5’ end of the mRNA molecule is capped at this site by the attachment of a specialized nucleotide (7-methyl guanosine).
The CAP site is followed by the initiation, or start codon (ATG), which specifies the start of translation; hence, according to the genetic code every polypeptide begins with methionine.
The DNA coding sequence for a gene in eukaryotes is not contiguous or uninterrupted. Each gene contains DNA sequences that code for the amino acid sequence of the protein, which are called exons.
These exons are interrupted by non-coding DNA sequences, which are called introns. The last exon ends with a stop codon (TAA, TAG or TGA), which represents the end of the gene-coding region and it is followed by the terminator sequence in the DNA sequence that defines the end of the gene-coding region. The 3 UTR of the mRNA molecule includes a poly(A) signal (AATAAA) that is added to the mRNA molecule following transcription.
Control of Gene Expression and Transcription
All individual cells of an organism contain the complete genetic blueprint of the organism. Therefore, it is of critical importance that the right genes are expressed in the correct tissue, at the proper level and at the correct time.
Temporal and tissue-specific gene expression in eukaryotic cells is mostly controlled at the level of transcription initiation. Two types of factors regulate gene expression: cis-acting control elements and trans-acting factors.
Cis-acting elements do not encode proteins; they influence gene transcription by acting as binding sites for proteins that regulate transcription. These DNA sequences are usually organized in clusters, located in the promoter region of the gene, that influence the transcription of genes. The TATA and CAAT boxes are examples of cis-acting elements.
The Trans-acting factors are known as transcription factors or DNA binding proteins. Transcription factors are proteins that are encoded for by other genes, and include steroid hormone receptor complexes, vitamin-receptor proteins and mineral-protein complexes. These transcription factors bind to specific DNA sequences in the promoter region of the target gene and promote gene transcription.
The precise mechanism(s) of how transcription factors influence gene transcription is not fully understood. However, transcription factors in general share some properties. They often have amino acid domains that contain one or more zinc ions, so-called zinc fingers that enable binding to DNA in a sequence-specific manner.
Another type of domain contains predominantly the amino acid leucine, the leucine zipper, and has a function in binding (‘zipping’) to similar domains in other proteins based on the charge of the individual amino acids.
Beside these domains, transcription factors often contain a nuclear localization signal to direct the protein from the cytoplasm to the nucleus and specific amino acids that can be modified by, for instance, phosphorylation, ubiquitinylation or acetylation and thereby get activated or inactivated. These modifications enable fine-tuning of the action of transcription factors.
As mentioned, transcription factors play a key role in temporal and tissue-specific gene expression. On binding they are able to influence the transcription of usually more than one gene.
There is ample evidence that transcription factors can remodel the chromatin structure in such a way that certain parts of the genome become available for transcription, while other parts of the chromosomes become tightly packed and inaccessible for transcription.
For example, the genes for the enzymes of gluconeogenesis can be turned on in the hepatocyte, but not in the adipocyte. This is because the hepatocyte expresses the transcription factors that are required to initiate the expression of the gluconeogenic enzymes.
Mastery regulatory proteins regulate the expression of many genes of a single metabolic pathway. For example, in the lipogenic pathway the fatty acid synthase complex codes for seven distinct genes that have to be coordinately expressed to form the enzymes required for fatty acid synthesis. This ensures that sufficient levels of all the enzymes of this metabolic pathway are available simultaneously.
- Alberts B, Bray D, Lewis J, Raff M, Roberts K, Watson JD. Molecular biology of the cell. 3rd ed. New York: Garland, 1994.
- Darnell J, Lodish H, Baltimore D. Molecular cell biology. 2nd ed. New York: Scientific American Books, WH Freeman, 1990.

